Snakemake introduction
What is snakemake?
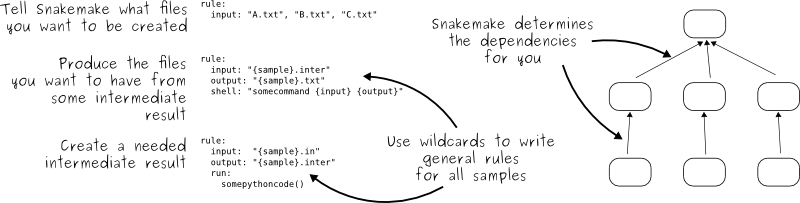
Snakemake is a workflow management and job scheduling tool which allows you to produce reproducible and scaleable bioinformatics workflows / pipelines. Workflow management tools allow you to automate multi-step bioinformatics analyses (e.g. data collection, QC, processing and visualisation) and organise complex pipelines in a human readable manner.
Why should I use snakemake?
- Automation: Allows you to automate mundane steps of bioinformatics process to focus more on data analysis
- Reproducibility: Promotes scientifc reproducibility
- Error tracking: Errors are easier to track reducing effort required to find and correct them
- Portability: Rule based system for subprocess organisation makes it easy to reuse / adapt workflow across projects / systems
- Readability: Snakemake uses python
- File creation is tracked:
- By completion: If a process crashes unexpectedly snakemake will automatically delete corrupted / incomplete files
- By date: so if you change a file at stage 2 of the process snakemake will infom you to rerun all subsequent processes
- Pipeline management and modularisation: Promotes the modularisation of bioinformatics processes into digestable chunks
Benefits of using snakemake on a computer cluster
- Makes parellisation simple: Manages scheduling of job submission to cluster (or to the cloud)
- Easily assign default, and / or subprocess specific, resources: No need for multiple shell scripts or SGE / SLURM headers i.e. parameters like
pe smpandh_vmemcan be specified in a snakemake profile - Supports all languages: Any type of script can be run, and shell and python commands can be executed directly
- Intermediate files can be automatically removed: temporary directory / file removal is simple
- Supports benchmarking: For example, to report CPU and memory usage
- Supports logging: control messages/errors
- Supports config files to abstract project specific details like filenames from the pipeline to promote code resuability and portability Supports use of environment modules: Environment modules on your local cluster can be pre-loaded in rule specific manner
- Supports Conda environments and package management
- Supports containers
- Many pre-written wrappers for common bioinformatics tasks: No need to reinvent the wheel
Alternatives to Snakemake
Move on to snakemake environment setup, or back to index page.